First of all, as a response to the first ToDo issue in the previous post, two different codes to calculate the order parameters from united atom simulations have been contributed and also thoroughly discussed (see discussion in the previous post and buildH program). This enabled the analysis of previously contributed united atom simulations and the results are now added into the manuscript. Also, significant amount of new data has been contributed and we have now a good collection of force fields for both PE and PG lipids.
The data is still suggesting that the CHARMM36 simulations capture the essential differences between PC, PE, PS, and PG headgroups. A figure of structural ensembles of different headgroups from CHARMM36 simulations is now contributed.
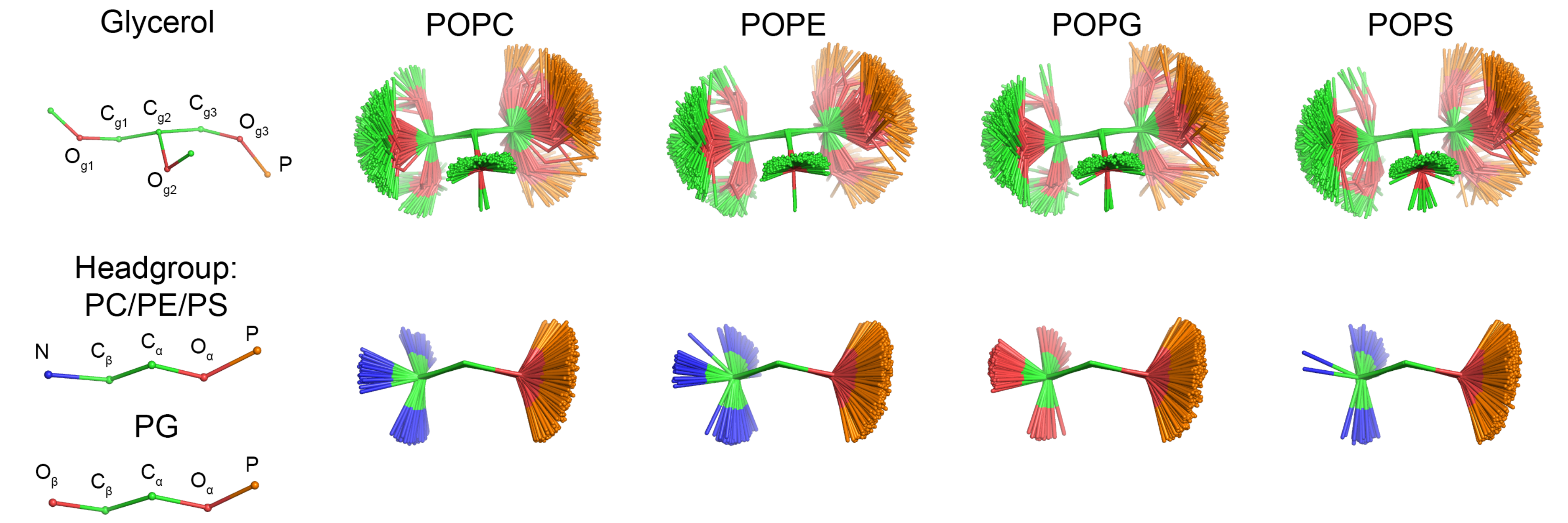 |
| Figure 1: Overlayed snapshots of a glycerol backbone and headgroup conformations of different headgroups from CHARMM36 simulations. Figure is made by Pavel Buslaev. |
For calcium-binding affinity to PG containing membranes we still need more data. The current data is not fully converged, and I think that we need longer CHARMM36 simulations with the recent NBfix correction for calcium and counterions. I have opened an issue in GitHub with more details for further discussion.
I have opened also other issues in GitHub. All the issues are important, but the most critical for the progress of the manuscript are the above mentioned analysis of structural differences between different lipid headgroups and additional data for PG-calcium interactions. In addition to the issues listed in GitHub, there are also Todo points in the manuscript (tex file) and SI (tex file). For example, we need citations for the force fields in tables I-III, and to finish the method sections for MD simulation and NMR experiments.