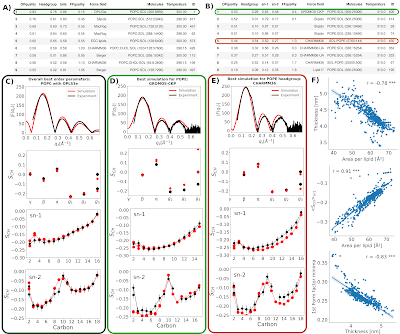
The NMRlipids III project was originally started to understand the discrepancies in form factors between simulations and experiments for cholesterol-containing membranes reported by Peter Heftberger et al. After experimental form factors for a series of POPC/cholesterol mixtures were reported by the same authors, the aim was to evaluate different simulations against experimental scattering and NMR data in order to find the parameters that would best capture lipid–cholesterol interactions.
Thanks to many contributors, a set of force fields is compared against experimental data in the current manuscript. However, the conclusions from the data are complicated because quality ranking of these simulations is not straightforward. Therefore the NMRlipids III project has been on hiatus for some time.
In the meanwhile, the Slipids model has been updated, Lipid17 and Slipids support has been added to CHARMM-GUI, and quality evaluation protocol has been quantified in the NMRlipids Databank. Because the popularity of Lipid17 and Slipids in cholesterol-containing membrane simulations is expected to increase due to the CHARMM-GUI support, we believe that the NMRlipids III project is still highly relevant and should be completed.
Moreover, in another project, Matti Javanainen analyzed lipid lateral diffusion coefficients in POPC/cholesterol mixtures simulated with CHARMM36. After applying the periodic-boundary-condition (PBC) correction by Vögele et al., the lateral diffusion coefficients agreed neither quantitatively nor qualitatively with those extracted from NMR experiments. Therefore, Matti extended the comparison to other force fields, namely Lipid17, Slipids-2020, and MacRog, and observed clear differences between their behavior. We believe that these data would nicely complement the structural evaluation against form factors and order parameters in the NMRlipids III project.
Current plan is that the NMRlipids III publication would contain the structural evaluation of POPC/cholesterol mixtures against scattering form factors and order parameters for the systems that are available from the NMRlipids Databank, and comparison of selected force fields against experimental lateral diffusion coefficients of lipids. New simulations with multiple cholesterol concentrations and box sizes, required for the PBC-correction, are already available in Zenodo for CHARMM36, Slipids-2020, Lipid17, and MacRog. These data will be soon added into the NMRlipids Databank. Also the diffusion analyses have already been performed.
We aim to draft the new version of the manuscript during autumn, after which we are looking for feedback from all contributors. Especially ideas on analyses that could explain the differences in diffusion/viscosity as a function of cholesterol between the different models would be helpful.
Because the analysis of lipid lateral diffusion coefficients will be a crucial part of the new manuscript, both Matti and Samuli will be corresponding authors, of the NMRlipids III publication. Otherwise the authorship policy remains as previously.