 |
| Figure 1: Headgroup and glycerol backbone order parameters of POPC without ions from Charmm-Drude and Charmm36 simulations |
 |
| Figure 2: Changes in the headgroup order parameters upon addition of ions in Charmm-Drude and Charmm36 simulations. |
 |
| Figure 1: Headgroup and glycerol backbone order parameters of POPC without ions from Charmm-Drude and Charmm36 simulations |
 |
| Figure 2: Changes in the headgroup order parameters upon addition of ions in Charmm-Drude and Charmm36 simulations. |
After the previous post on the NMRlipids IVb manuscript, I have made further analysis on the structural differences between PC, PE, PG and PS headgroups using the development version of the NMRlipids databank and CHARMM36 simulations, which give the headgroup and glycerol backbone order parameters closest to experiments for all lipids. Based on these analyses, I have reformulated the NMRlipids IVb manuscript to focus on the headgroup conformations of different lipids.
Main points of the manuscript are now:
1) Differences in NMR order parameters between different headgroups can be explained by the changes in dihedral angle distributions, suggesting that similar conformations are accessed by all headgroups, but with different probabilities.
CHARMM36 simulations approximately reproduce the main differences of headgroup order parameters between PC, PE, PG and PS lipids (forking of the ⍺-carbon in PS and positive β-carbon value in PG in Figs. 1 A and B). These differences can be explained by the differences in the heavy atom dihedral distributions (Fig. 1 C).
2) The changes in headgroup order parameters upon addition of charged molecules can be explained by the changes in dihedral angle distributions close to the phosphate region.
CHARMM36 simulations reproduce the experimental changes in headgroup order parameters upon addition of cationic surfactants which are related to the substantial tilt of P-N dipole (Fig. 2 A) and changes in dihedral distributions close to the phosphate region (Fig. 2 B).
 |
| Figure 2: Response of POPC headgroup A) order parameters, P-N vector and B) dihedral angles to the addition of cationic surfactant from CHARMM36 simulations compared with experimental data. |
3) Wide range of headgroup conformations are observed also in lipids that are directly bound to proteins, suggesting that the specific binding of lipids to proteins is dominated by the intermolecular lipid-protein interactions rather than the differences in conformational restrictions of lipids.
Dihedral angles calculated from protein bound lipid structures from the protein data bank (PDB) have wide distributions independently on the headgroup chemistry (Fig. 3).
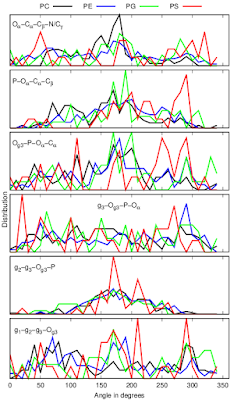 |
| Figure 3: Dihedral distributions of protein bound lipids analysed from the protein data bank (PDB). |
Comparison of headgroup and glycerol backbone order parameters between different force fields and experiments, and results from lipid mixtures are now in the supplementary information because conclusions from these are essentially the same as in NMRlipids I and IV publications. None of the force fields correctly capture the conformational ensembles of lipid headgroups, but CHARMM36 is closest, and simulations containing charged lipids are complicated by the overestimated binding affinity of counterions.
I believe that the manuscript is approaching the submission stage. Because MD simulations from the NMRlipids project are able to explain the differences in experimental order parameters between different lipids, thereby giving an interpretation of the lipid conformational ensembles, my current plan is to submit the manuscript to the ACS Central Science.
Most critical things to do before submission are related to the methods, see todo points in the manuscript and especially in the supplementary information. There are also some issues still open in the GitHub. All kind of comments on the manuscript are welcomed.
 |
| Figure 1: Schematic structure of the new databank. Beta versions of the Databank, Databank builder and Databank analyzer codes are available. |