After the previous post on the NMRlipids IVb manuscript, I have made further analysis on the structural differences between PC, PE, PG and PS headgroups using the development version of the NMRlipids databank and CHARMM36 simulations, which give the headgroup and glycerol backbone order parameters closest to experiments for all lipids. Based on these analyses, I have reformulated the NMRlipids IVb manuscript to focus on the headgroup conformations of different lipids.
Main points of the manuscript are now:
1) Differences in NMR order parameters between different headgroups can be explained by the changes in dihedral angle distributions, suggesting that similar conformations are accessed by all headgroups, but with different probabilities.
CHARMM36 simulations approximately reproduce the main differences of headgroup order parameters between PC, PE, PG and PS lipids (forking of the ⍺-carbon in PS and positive β-carbon value in PG in Figs. 1 A and B). These differences can be explained by the differences in the heavy atom dihedral distributions (Fig. 1 C).
2) The changes in headgroup order parameters upon addition of charged molecules can be explained by the changes in dihedral angle distributions close to the phosphate region.
CHARMM36 simulations reproduce the experimental changes in headgroup order parameters upon addition of cationic surfactants which are related to the substantial tilt of P-N dipole (Fig. 2 A) and changes in dihedral distributions close to the phosphate region (Fig. 2 B).
 |
| Figure 2: Response of POPC headgroup A) order parameters, P-N vector and B) dihedral angles to the addition of cationic surfactant from CHARMM36 simulations compared with experimental data. |
3) Wide range of headgroup conformations are observed also in lipids that are directly bound to proteins, suggesting that the specific binding of lipids to proteins is dominated by the intermolecular lipid-protein interactions rather than the differences in conformational restrictions of lipids.
Dihedral angles calculated from protein bound lipid structures from the protein data bank (PDB) have wide distributions independently on the headgroup chemistry (Fig. 3).
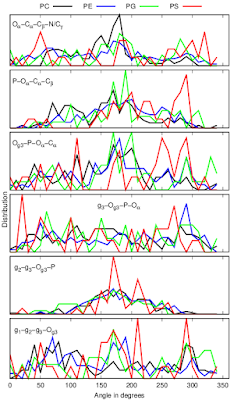 |
| Figure 3: Dihedral distributions of protein bound lipids analysed from the protein data bank (PDB). |
Comparison of headgroup and glycerol backbone order parameters between different force fields and experiments, and results from lipid mixtures are now in the supplementary information because conclusions from these are essentially the same as in NMRlipids I and IV publications. None of the force fields correctly capture the conformational ensembles of lipid headgroups, but CHARMM36 is closest, and simulations containing charged lipids are complicated by the overestimated binding affinity of counterions.
I believe that the manuscript is approaching the submission stage. Because MD simulations from the NMRlipids project are able to explain the differences in experimental order parameters between different lipids, thereby giving an interpretation of the lipid conformational ensembles, my current plan is to submit the manuscript to the ACS Central Science.
Most critical things to do before submission are related to the methods, see todo points in the manuscript and especially in the supplementary information. There are also some issues still open in the GitHub. All kind of comments on the manuscript are welcomed.

An online meeting on finalizing the NMRlipids IVb manuscript will take place on Tuesday 12th of January starting at 11.00 AM CET in Zoom.
ReplyDeleteIf you are interested to join, send me email to get the meeting ID.
Here is the list of topics discussed in the meeting last week and some actions made based on those.
Delete1) We decided to keep the layout of figure 2 (at least for now). The polished figure is now updated in the manuscript. There are some further comments in the comments below and in Overleaf.
2) Figure showing the comparison of ion binding affinity to PG containing membranes between different simulations and experiments is now moved to SI. The text has been modified accordingly, but may be still too technical.
3) Markus Miettinen agreed to polish figures S4 and S5, also discussed in issues https://github.com/NMRLipids/NMRlipidsIVPEandPG/issues/5 and https://github.com/NMRLipids/NMRlipidsIVPEandPG/issues/1
4) Enantiomers in PG headgroups have now been analyzed from simulations and manuscript updated accordingly, see issue https://github.com/NMRLipids/NMRlipidsIVPEandPG/issues/38
5) An issue about the title for the manuscript has been opened: https://github.com/NMRLipids/NMRlipidsIVPEandPG/issues/39
6) Differences in PS dihedrals next to phosphate were considered negligible in previous version. However, as pointed out by Markus Miettinen, similar changes cause order parameter decrease upon addition of charged molecules in figure 3. The text is now slightly updated to be more consistent.
7) We discussed if it would be possible to analyze membrane bound lipid conformations from GPCRMD project (gpcrmd.org). This is yet to be done.
8) More coarse presentation (larger widths for angle windows) of protein bound lipid dihedrals in figure 4 was suggested. Also figures showing different lipids bound to proteins in similar conformations were suggested. After the meeting, an issue about the quality of lipid structures in PDB was opened (https://github.com/NMRLipids/NMRlipidsIVPEandPG/issues/40). All these three issues are more or less related and I have a plan to start to look at this asap.
9) Contributors were reminded on the importance of writing the simulation details in the designated locations in the SI.
10) In addition to these topics, urgent matters are the finalizing the experimental and simulation method sections, as well as figures showing experimental data (Figs. 1, and S1-S3). Dihedral figures in SI are now polished.
Please comment if something is missing.
Regarding point (2), I have now updated the MD methods and results and discussion, and moved the most force field comparison to the SI.
DeleteRegarding point (7), it seems that there does not currently exist a straightforward way to automatically analyze the data in GPCRMD project. Nevertherless, it might be reasonable to check if there is data from same proteins which we already have in the PDB analysis and analyze these manually.
I checked the overlapping proteins in our analysis and in GPCRmd and found one: PDBid 4k5y. However, this protein has a PG lipid attached in PDB, but it is simulated in pure POPC bilayer in GPCRmd (as almost all the simulations in GPCRmd). Therefore it is not useful for our current purpose.
DeleteTo ease the commenting and updating the manuscript for contributors who are less familiar with the Git, we have now added the manuscript in the Overleaf (www.overleaf.com).
ReplyDeleteThe overleaf project is also connected to the GitHub repository of the project (https://github.com/NMRLipids/NMRlipidsIVPEandPG) through the submodule in the Manuscript folder. Therefore, if you prefer modifying locally and commit to GitHub, you can continue doing this by taking into account that the Manuscript folder is now a submodule and will be kept up-to-date with the overleaf.
If you use overleaf, you do not have to care about the Git unless you want to. We will take care of the commits between Git and overleaf.
Let me know if you have questions or comments on this, or if you wish to have access to the Overleaf project. Aim of this exercise is to ease the communication about manuscripts utilizing the Overleaf because not everyone are familiar with the Git.
It might be more intuitive for the reader, if we would show the dihedral populations as free energies (in units of kT) instead of probabilities?
ReplyDeleteDo you mean only the populations in Fig.2 A), or also dihedral distributions?
DeletePersonally I find probabilities and densities more easier concepts than energy, but maybe for populations in Fig. 2 A) this might be a good idea.
I was thinking of the dihedral distributions in 2C, 3B, and 4. I like free energy (in units of kT) rather intuitive, but I this is of course a matter of taste.
DeleteThe probabilities often go to 0 (dG=inf), thus making the plots harder to read. So I'd stick to probabilities. P.s. the Manuscript folder has a funny name in GitHub now, so the links in this blog post don't work.
DeleteThe dG=inf point is true, meaning that we would need to cut the y-axis. However, it would also mean that these (P=0) regions would better stick out from the plot, which would be useful, because some of our main points are about such regions existing.
DeleteI will try and fix the links now.
I think that this may not make sense for figure 4 because the probabilities come from different systems.
DeleteI have now made figures in relative kT units using -log(distribution) and setting the minimum value of the y-axis in zero in each system. Figures are here:
Deletehttps://github.com/NMRLipids/NMRlipidsIVPEandPG/blob/master/Figs/DIHEDRALSlog.pdf
https://github.com/NMRLipids/NMRlipidsIVPEandPG/blob/master/Figs/HGorderparametersPCvsSURFchangeDIHEDRALSlog.pdf
I think that the plots reveal that the density was not exactly zero in some regions which looked zero in the distribution plot. Also, there are not many regions where distribution is exactly zero (energy=inf). I have cutted out these regions from the plot by tuning the y-axis range. Therefore, I think now that we should use these plots instead of the distributions.
I think that simple -log(distribution) plot does not make sense in Figure 4, because the individual points are from different systems. Instead, I made a some analysis using the kT plots above. For each dihedral angle observed in PDB, I took the energy value (in kT units) from the plot from simulation. Then I calculated the normalized distribution of energies for each dihedral in different lipids observed in PDB:
https://github.com/NMRLipids/NMRlipidsIVPEandPG/blob/master/Figs/ENEDISTfromPDB.pdf
I think that this plot gives us some impression on the energy cost of individual dihedrals upon binding to proteins. We have to be careful in using correct terms when discussing these, but I think that we should consider showing also this kind of plot in the main article.
I have now updated the dihedral distributions in figures 2 and 3 to -log(distribution) scale and modified the text accordingly.
DeleteIn figure 4, there are still dihedral distributions because the data there comes from different systems with different environments, and I think that the link from distributions to energies is too complicated in this case.